ORIGINAL ARTICLE
Genomic Surveillance of SARS-CoV-2 Variants at a Reference Cancer Hospital in Rio de Janeiro, Brazil
Vigilância Genômica de Variantes do SARS-CoV-2 em um Hospital Referência para o Tratamento de Câncer no Rio de Janeiro, Brasil
Vigilancia Genómica de Variantes del SARS-CoV-2 en un Hospital de Referencia para el Tratamiento del Cáncer en Río de Janeiro, Brasil
https://doi.org/10.32635/2176-9745.RBC.2024v70n3.4637
Élida Mendes de Oliveira1; Caroline Carvalho de Sá2; Julia Botto de Barros Cordeiro3; Luiz Claudio Santos Thuler4; Maria Eduarda Lanzillota Assumpção5; Gabriella Seara de Andrade6; Vinicius Figueiredo Vizzoni7; João Paulo de Biaso Viola8; Marcelo Alves Soares9; Juliana Domett Siqueira10; Livia Ramos Goes11
1-3,5,6,9-11Instituto Nacional de Câncer (INCA), Programa de Genética e Virologia Tumoral. Rio de Janeiro (RJ), Brasil. E-mails: mendeselida16@gmail.com; caroline.sa@edu.unirio.br; jubbcordeiro@gmail.com; lanzieduarda@edu.unirio.br; searagabriella@gmail.com; masoares@inca.gov.br; sidoju.juliana@gmail.com; livia.goes@inca.gov.br. Orcid iD: https://orcid.org/0009-0005-1086-9652; Orcid iD: https://orcid.org/0009-0007-9337-7956; Orcid iD: https://orcid.org/0009-0005-2199-6534; Orcid iD: https://orcid.org/0009-0008-4315-3601; Orcid iD: https://orcid.org/0000-0003-0186-986X; Orcid iD: https://orcid.org/0000-0002-9013-2570; Orcid iD: https://orcid.org/0000-0002-4266-9795; Orcid iD: https://orcid.org/0000-0002-4909-4912
4INCA, Programa de Epidemiologia Clínica. Rio de Janeiro (RJ), Brasil. E-mail: lthuler@inca.gov.br. Orcid iD: https://orcid.org/0000-0003-2550-6537
7Instituto de Pesquisas Biomédicas, Laboratório de Biologia Molecular, Hospital Naval Marcílio Dias. Rio de Janeiro (RJ), Brasil. E-mail: viniciusfv@gmail.com. Orcid iD: https://orcid.org/0000-0002-7595-1607
8INCA, Programa de Imunologia e Biologia Tumoral. Rio de Janeiro (RJ), Brasil. E-mail: jpviola@inca.gov.br. Orcid iD: https://orcid.org/0000-0002-0698-3146
Corresponding authors: Livia Ramos Goes; Juliana Domett Siqueira. Rua André Cavalcanti, 37, 4° andar – Centro. Rio de Janeiro (RJ), Brasil. E-mails: liviargoes@gmail.com; sidoju.juliana@gmail.com
ABSTRACT
Introduction: The fast SARS-CoV-2 spread and high mutation rates during viral replication led to virus diversification and the emergence of new variants. Genomic surveillance has been key to monitoring SARS-CoV-2 variants across the globe. Immune suppression, as observed in cancer patients, is a risk factor for SARS-CoV-2 infection and severe COVID-19. Objective: To report a two-year genomic surveillance of SARS-CoV-2 in cancer patients followed up at the Brazilian National Cancer Institute, Rio de Janeiro, Brazil. Method: Prospective observational study with 384 SARS-CoV-2+ swabs specimens collected between October 2020 and September 2022. SARS-CoV-2 spike was analyzed by PCR and Sanger sequencing to determine the infecting variant. Results: Most of the patients had solid organ malignancies (298/384; 77.6%) and 16.1% (62/384) had metastatic disease. Severe COVID-19 cases accounted for 29.4% (113/384) and 27.1% (104/384) of deaths registered. The most common SARS-CoV-2 infecting variants were Gamma (n=137) and Omicron (BA.1) (n=73). The variant distribution overtime was similar to what has been reported for the general population of Brazil in the same period. When patients’ cancer topographies were analyzed, it was found that Gamma infected patients with breast (47/137; 34.3%) and cervical (11/137; 8%) cancer were more frequent than other variants, while Omicron predominated among rectum (10/122; 8.2%) and prostate (8/122; 6.6%) cancer compared to other variants. Conclusion: Genomic surveillance is an important tool for identifying and evaluating the impact of SARS-CoV-2 variants, and should continue especially in immunosuppressed populations.
Key words: SARS-CoV-2; COVID-19; Neoplasms/genetics; Epidemiological Monitoring; Genome, Viral.
RESUMO
Introdução: A rápida propagação do SARS-CoV-2 e as altas taxas de mutação durante a replicação viral levam à diversificação do vírus e ao surgimento de novas variantes. A vigilância genômica tem sido fundamental para monitorar as variantes do SARS-CoV-2 em todo o mundo. A supressão imunológica, conforme observada em pacientes com câncer, é um fator de risco para infecção por SARS-CoV-2 e covid-19 grave. Objetivo: Relatar uma vigilância genômica de dois anos do SARS-CoV-2 em pacientes com câncer acompanhados no Instituto Nacional de Câncer, Rio de Janeiro, Brasil. Método: Estudo observacional prospectivo. Foi avaliado um total de 384 amostras de swabs SARS-CoV-2+ coletadas entre outubro de 2020 e setembro de 2022. O pico do SARS-CoV-2 foi analisado por PCR e sequenciamento Sanger para determinar a variante infecciosa. Resultados: A maioria dos pacientes apresentava malignidades de órgãos sólidos (298/384; 77,6%) e 16,1% (62/384), doença metastática. Os casos graves de covid-19 representaram 29,4% (113/384) e foram registrados 27,1% (104/384) óbitos. Em relação às variantes infectantes do SARS-CoV-2, as mais comuns foram Gamma (n=137) e Ômicron (BA.1) (n=73). A distribuição da variante ao longo do tempo foi semelhante ao que foi relatado para a população geral do Brasil no mesmo período. Quando analisadas as topografias de câncer dos pacientes, a Gamma infectou pacientes com câncer de mama (47/137; 34,3%) e cervical (11/137; 8%), mais frequentemente do que outras variantes, enquanto a Ômicron predominou entre reto (10/122; 8,2%) e câncer de próstata (8/122; 6,6%) em comparação com outras variantes. Conclusão: A vigilância genômica é uma ferramenta importante para identificar e avaliar o impacto das variantes do SARS-CoV-2, e deve prosseguir especialmente em populações imunossuprimidas.
Palavras-chave: SARS-CoV-2; COVID-19; Neoplasias/genética; Epidemiological Monitoring; Genoma Viral.
RESUMEN
Introducción: La rápida propagación del SARS-CoV-2 y las altas tasas de mutación durante la replicación viral conducen a la diversificación del virus y la aparición de nuevas variantes. La vigilancia genómica ha sido clave para monitorear las variantes del SARS-CoV-2 en todo el mundo. La inmunosupresión, como se observa en pacientes con cáncer, es un factor de riesgo de infección por SARS-CoV-2 y COVID-19 grave. Objetivo: Informar una vigilancia genómica de dos años del SARS-CoV-2 en pacientes con cáncer seguida en el Instituto Nacional de Cáncer, Río de Janeiro, Brasil. Método: Estudio observacional prospectivo. Se evaluaron un total de 384 muestras de hisopos de SARS-CoV-2+ recolectadas entre octubre de 2020 y septiembre de 2022. El pico de SARS-CoV-2 se analizó mediante PCR y secuenciación Sanger para determinar la variante infectante. Resultados: La mayoría de los pacientes tenían neoplasias malignas de órganos sólidos (298/384; 77,6%) y el 16,1% (62/384), enfermedad metastásica. Los casos graves de COVID-19 representaron el 29,4% (113/384) y se registraron el 27,1% (104/384) de las muertes. En cuanto a las variantes infectantes del SARS-CoV-2, las más comunes fueron Gamma (n=137) y Ómicron (BA.1) (n=73). La distribución de variantes en el tiempo fue similar a lo reportado para la población general del Brasil en el mismo período. Cuando se analizaron las topografías de cáncer de los pacientes, la Gamma infectó a los pacientes con cáncer de mama (47/137; 34,3%) y de cuello uterino (11/137; 8%) con mayor frecuencia que otras variantes, mientras que Ómicron predominó entre el recto (10/122; 8,2%) y cáncer de próstata (8/122; 6,6%) en comparación con otras variantes. Conclusión: La vigilancia genómica es una herramienta importante para identificar y evaluar el impacto de las variantes del SARS-CoV-2 y debe continuarse, especialmente en poblaciones inmunodeprimidas.
Palabras clave: SARS-CoV-2; COVID-19; Neoplasias/genética; Monitoreo Epidemiológico; Genoma Viral.
INTRODUCTION
The emergence of SARS-CoV-2 infections causing COVID-19, posed a risk to public health worldwide in the last years. COVID-19 was declared a pandemic by the World Health Organization (WHO) in March 20201.The rapid transmission of SARS-CoV-2 across the globe and the high mutation rates characteristic of RNA viruses accelerated the diversification of the virus during the pandemic. In this regard, genomic surveillance has been essential to detect the emergence of new SARS-CoV-2 variants. The US Centers for Diseases Control (CDC) has proposed a classification system based on the pathogenic characteristics of different SARS-CoV-2 variants. Variants of concern (VOC) are viral strains which harbor genomic changes that have been proven to modulate biological features such as increased transmissibility or the affinity to the ACE-2 receptor2.
Variants of interest (VOI) are lineages that contain mutations known to alter viral fitness leading to more severe cases or affecting diagnosis, and which are closely monitored for their potential to change viral dynamics. This system is continually updated to reflect the scenario of circulating variants. Since the beginning of the pandemic, six VOCs have been identified: Alpha (B.1.1.7 and Q lineages), Beta (B.1.351 and descendent lineages), Gamma (P.1 and descendent lineages), Delta (B.1.617. 2 and descendent lineages), Epsilon (B.1.427 and B.1.429) and Omicron (B.1.1.529 and descendent lineages). Currently, the unique variant considered a VOC is Omicron, and the remaining were downgraded to variants being monitored (VBM) due to their low or non-existent circulation3.
At the onset of the pandemic, the lineages circulating in Brazil were identified as B.1.1, B.1.1.28, and B.1.33. In October 2020, a VOI known as P.2 emerged and gained prominence in subsequent months, becoming the most prevalent lineage in the country. Then, the Gamma variant emerged, initially detected by genomic sequencing in December 2020. The occurrence of this variant was associated with an increase in deaths during 20214 due to its ability to spread faster than the variants circulating at that time, and acquiring mutations in the Spike protein such as E484K and N501Y that have been associated with immune evasion and higher transmissibility5.
The Gamma variant was then replaced by Delta, which contained new mutations in the receptor binding domain (RBD) of the Spike protein and was responsible for new records of infections around the world, evading the humoral neutralization by antibodies induced either by previous infections or by vaccination6. At the end of 2021, the Omicron variant was detected in South Africa and spread rapidly across the world, becoming the most prevalent in several countries, including Brazil7. Over time, subvariants of Omicron emerged such as BA.1, BA.2, BA.3, BA.4, BA.5, and since 2022, this variant is responsible for more than 98% of cases of infection by SARS-CoV-2 in Brazil8.
SARS-CoV-2 infections of immunosuppressed individuals, such as cancer patients, are considered a risk factor for unfavorable outcomes. The development of COVID-19 vaccines reduced significantly the risks of death or hospitalization. However, titers of seroconversion in patients with solid or hematological cancers are lower when compared to immunocompetent individuals9. It has also been shown that cancer patients have a higher intrahost SARS-CoV-2 diversity10 and can accumulate a handful of mutations during persistent infections, with potential links to the emergence of new strains as these genomic changes are established and transmitted11.
Therefore, the genomic surveillance of SARS-CoV-2 in this particular group is essential for tracking the virus evolution and understanding the mechanisms that lead to immune evasion and increased transmission. The data presented herein show the genomic surveillance of SARS-CoV-2 infections over a two-year period in cancer patients followed at the Brazilian National Cancer Institute (INCA), Rio de Janeiro, Brazil.
METHOD
This is a prospective observational study in which all cancer patients registered at the INCA, Rio de Janeiro, Brazil, diagnosed with COVID-19 in the period between October 2020 and September 2022 were eligible for participation. SARS-CoV-2 infection was diagnosed through naso-and oropharyngeal swab specimens by real-time reverse-transcription polymerase chain reaction (RT-qPCR). Up to forty samples with cycle threshold (Ct) < 30 at the diagnostic RT-qPCR were selected per each month of the study, whenever available. Twenty consecutive samples were selected every two weeks when it was possible or completed the 40 samples with the following two-week period of the same month otherwise. In order to avoid cases of patients with consecutive SARS-Cov-2 positive tests or reinfection, only the first episode (test) was included in the study.
While the month of September 2022 had only 2 samples and both failed to be PCR-amplified, SARS-COV-2 diagnosis was not performed in March and April 2022, a period where samples were tested at another diagnostic center in Rio de Janeiro (outside INCA), and therefore no samples were available during that bimester. Selected samples were submitted to PCR reactions as described below and all samples successfully amplified were included in the study (n = 384). Viral RNA was extracted from the residual material used for COVID-19 diagnosis with the QIAamp MiniElute Virus Spin Kit (QIAGEN, Chatsworth, CA, USA) according to the manufacturer’s instructions. Subsequently, cDNA was synthetized using the SuperScriptTM III First-Strand Synthesis System (Thermo Fisher Scientific, Waltham, MA, USA).
A SARS-CoV-2 S gene fragment of 1,532-bp (spanning amino acid positions 320-760, based on Wuhan-Hu-1 SARS-CoV-2 S gene Genbank acc# MN90894) was amplified by PCR using the primers 74_LEFT and 78_RIGHT from the ARTIC network nCov-2019 V.3 primer set (Artic Network)12 according to the protocol described by Goes13. This DNA fragment comprises the main mutation sites to the different variants that circulated in Brazil during the study period.
An additional PCR was performed to distinguish P.2 from N.9 variants. An ORF1 fragment of 1,032-pb (corresponding to amino acid positions 3367-3709, based on Wuhan-Hu-1 SARS-CoV-2 S gene) was PCR-amplified with the primers 35_LEFT and 37_RIGHT (Artic Network)2. The PCR conditions were 95ºC for 5 min, 40 cycles of 95ºC for 15 s, 60ºC for 30 s and 72ºC for 1 min, followed by a final extension at 72ºC for 5 min.
PCR products were purified with the ReliaPrep™ DNA Clean-Up and Concentration System kit (Promega, Madison, WI) and sequenced by Sanger methodology. The BigDyeTM Terminator v3.1 Cycle Sequencing kit (Thermo Fisher Scientific, Waltham, MA) was used for sequencing reactions with the following PCR primers: 74_LEFT, 76_LEFT, 76_RIGHT, 77_LEFT, 77_RIGHT and 78_RIGHT for the S gene fragment and 35_LEFT and 37_RIGHT for the ORF1 gene fragment. Nucleic acid sequences were determined using an automated ABI 3130xl Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA). Electropherograms were assembled to the Wuhan-Hu-1 SARS-CoV-2 reference sequence (Genbank acc# MN908947) and manually edited with SeqMan (DNAStar, inc. Madison, WI).
SARS-CoV-2 S fragment sequences obtained were aligned with sequences extracted from the GISAID EpiCoV database representing different PANGO lineages that circulated in Brazil during the study period. Maximum likelihood phylogenetic reconstruction was performed with IQ-TREE v1.5.514 using this alignment and the nucleotide substitution model assessed with ModelFinder15. The approximate likelihood-ratio test (aLRT) provided clade support. ORF1 sequences were compared to the Wuhan-Hu-1 SARS-CoV-2 reference sequence (Genbank acc# MN908947) to identify changes L3468V and F3605L, characteristic of P.2 and N.9, respectively.
COVID-19 severity was defined as follows: mild – no requirement of oxygen support, moderate – use of non-invasive oxygen support, severe – mechanical ventilation or death. The overall distribution between cases of cancer according to topography and infecting SARS-CoV-2 variants had its deviation from randomness estimated by a Chi-square test with 32 degrees of freedom (nine topographies versus five viral variants). Specific associations between viral lineages and cancer topographies were estimated with Pearson`s chi-square tests comparing each lineage with all the remaining in pairwise comparisons.
The National Ethics Committee (CONEP) (CAAE (submission for ethical review): 30608220.8.0000.5274) approved the study, report number 4152775, in compliance with Directive 466/201216 of the National Health Council for studies involving human beings and according to Good Clinical Practices in regard to the patients confidentiality.
RESULTS
The median age of the patients investigated was 59 years, mostly females (64.6%). Solid organ malignancies were the most prevalent (77.6%) and 16.1% metastasized. The predominant type of cancer in the series was breast cancer (21.6%). Severe cases of COVID-19 were diagnosed in 29.4% of the patients and 27.1% deaths occurred. The main COVID-19 symptoms reported by the patients were fever (16.1%), cough (15.9%), dyspnea (10.7%), coryza (3.1%) and headache (0.3%) (Table 1).
Table 1. Demographic and clinic characteristics of the 384 cancer patients included in the study
|
Characteristic |
Patients (%) |
|
Age (years) |
|
|
< 25 years |
56 (14.6) |
|
25-64 years |
204 (53.1) |
|
≥ 65 years |
124 (32.3) |
|
Sex |
|
|
Male |
136 (35.4) |
|
Female |
248 (64.6) |
|
COVID-19 severity |
|
|
Mild |
206 (53.6) |
|
Moderate |
65 (16.9) |
|
Severe |
113 (29.4) |
|
Deaths |
|
|
COVID-19-related |
104 (27.1) |
|
Other causes |
29 (7.5) |
|
Primary cancer |
|
|
Solid Tumor |
298 (77.6) |
|
Hematological |
86 (22.4) |
|
Metastatic cancer |
62 (16.1) |
The SARS-CoV-2 infecting variants found were classified according to the phylogenetic analysis. The ancestral lineages B.1, B.1.1, B.1.1.28, B.1.1.33 and the N.1 were grouped since they could not be distinguished considering the genomic region studied. Figure 1 shows a representative phylogenetic tree used for variant classification.
Most of the patients (99.2%) had their SARS-CoV-2 variant successfully determined. Among the analyzed variants, 35.7% of the patients were infected by Gamma (P.1) and 19% had the Omicron (BA.1), as shown in Table 2. All samples classified as P.2/N.9 by S gene phylogeny were identified as P.2 using the ORF1 region. Three samples (0.8%) had discordant results between S (classified as P.2/N.9) and ORF1 (classified as B.1) regions and remained unclassified.
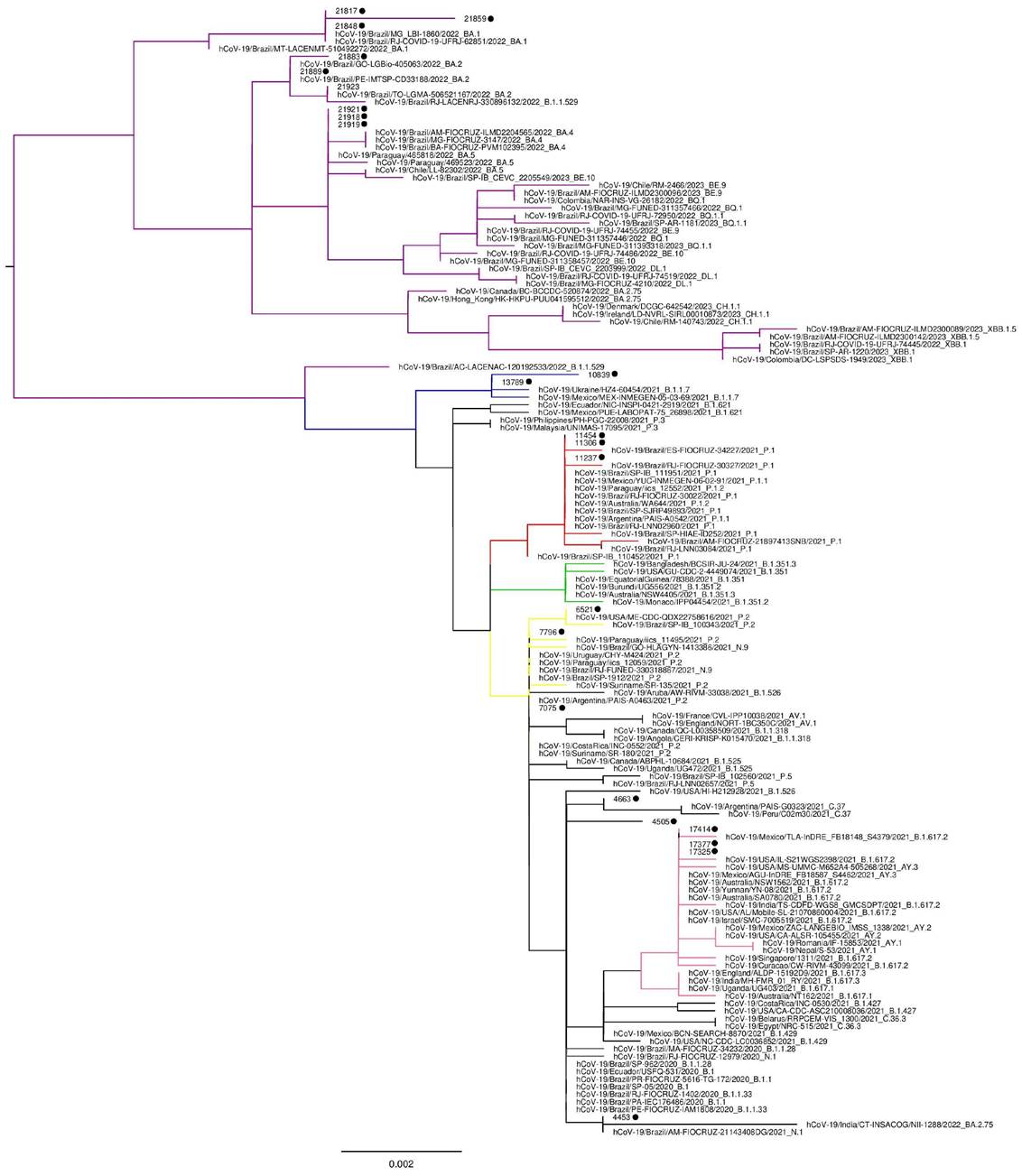
Figure 1. Representative phylogenetic tree showing SARS-CoV-2 variant classification. Omicron variants are colored in purple, Alpha in blue, Gamma in red, Beta in green, P.2 and N.9 in yellow and Delta in light pink. Sequences generated in this study are shown with a black dot for easy visualization
Table 2. Distribution of SARS-CoV-2 variants among the patients
|
Variant |
N (%) |
|
B.1/B.1.1/B.1.1.28/B.1.1.33/N.1 |
20 (5.2) |
|
Alpha (B.1.1.7) |
2 (0.5) |
|
Delta (B.1.617.2/AY.3) |
49 (12.8) |
|
Gamma (P.1) |
137 (35.6) |
|
P.2 |
51 (13.3) |
|
Omicron (BA.1) |
73 (19.0) |
|
Omicron (BA.2) |
5 (1.3) |
|
Omicron (BA.4/5) |
44 (11.5) |
|
Indeterminate |
3 (0.8) |
SARS-CoV-2 variant distribution was also analyzed overtime as shown in Figure 2A. In October 2020, the ancestral lineages predominated and in the following months, P.2, a VOI at that time, became dominant. Gamma (P.1), the third variant classified as a VOC, started to grow rapidly in January 2021 and prevailed for the following months. In that period, the first VOC detected in the world, Alpha (B.1.1.7), was detected occasionally. Delta (B.1.617/AY.3) was detected in 12.8% samples in total and was the most frequent in the second semester of 2021. The month of October 2021 had only one sample and the following two months had only two samples each. In the beginning of 2022, the Omicron variant became dominant, more specifically the sublineage BA.1. The month of January 2022 was the richest in sequences, 64 in total, all classified as of BA.1 lineage.
Throughout the months of February and March 2022, and in the last month of the study (September 2022), there were no samples sequenced. The BA.2 sublineage was first detected in the surveillance in April 2022, as well as the first Omicron lineage BA.4/5 sequences. The latter was the prominent in the final four months of the study. In comparison with SARS-CoV-2 variant data reported by the Oswaldo Cruz Foundation (Fiocruz) in the general population of the country for the same period, no differences in variants´ epidemiological profiles were observed between both groups.
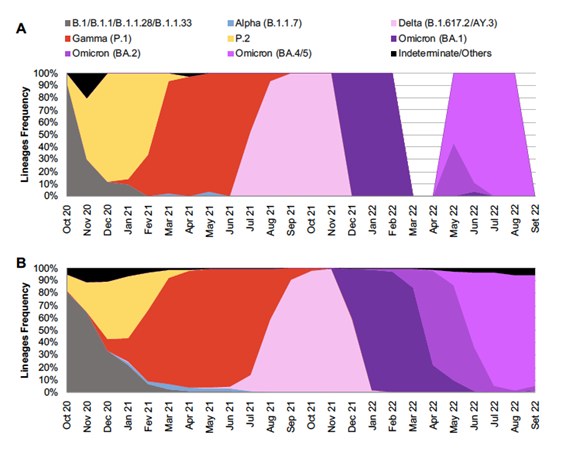
Figure 2. Distribution and frequency of SARS-CoV-2 variants found over time between October 2020 and September 2022 infecting patients at the Brazilian National Cancer Institute (A) and comparison with data from Brazil as reported by the Oswaldo Cruz Foundation (Fiocruz)
Source: The Authors, plus Dashboard Genomic Network data from Fiocruz17.
The distribution of patients' cancer topographies and their infecting SARS-CoV-2 variants is shown in Table 3. The association between viral variants and hematological and solid tumors were tested, but no differences have been found (data not shown). Cases of SARS-CoV-2 infection according to cancer topography varied among the different lineages. While analyzing this distribution, it has been noticed that Gamma infected patients with breast and cervical cancer more frequently than other variants did (p < 0.001) and Omicron predominated among rectum and prostate cancer patients compared to other variants (p <0.001). Ancestral lineages (B.1/B.1.1/B.1.1.28/B.1.1.33/N.1) and Delta did not show preponderance in any cancer topography over the remaining variants.
Table 3. Distribution of SARS-CoV-2 variants across different cancer topographies, INCA, October 2020 to September 2022
|
Topography |
B1 N (%) |
P2/N9 N (%) |
Gamma N (%) |
Delta N (%) |
Omicron N (%) |
Total |
|
|
Breast |
5 (25) |
18 (35.3) |
47 (34.3) |
10 (20.4) |
2 (1.6) |
82 |
|
|
Leukemia |
3 (15) |
7 (13.7) |
6 (4.4) |
7 (14.3) |
13 (10.7) |
36 |
|
|
Lymphoma |
2 (10) |
2 (3.9) |
13 (9.5) |
5 (10.2) |
22 (18) |
44 |
|
|
Cervical |
2 (1) |
0 (0) |
11 (8) |
3 (6.1) |
1 (0.8) |
17 |
|
|
Skin |
1 (5) |
3 (5.9) |
3 (2.2) |
0 (0) |
2 (1.6) |
9 |
|
|
Prostate |
0 (0) |
3 (5.9) |
3 (2.2) |
2 (4.1) |
8 (6.6) |
16 |
|
|
Lung |
0 (0) |
3 (5.9) |
9 (6.6) |
2 (4.1) |
8 (6.6) |
22 |
|
|
Rectum |
1 (5) |
1 (2) |
3 (2.2) |
2 (4.1) |
10 (8.2) |
17 |
|
|
Other |
6 (30) |
14 (27.5) |
42 (30.7) |
18 (36.7) |
56 (45.9) |
136 |
|
|
Total |
20 |
51 |
137 |
49 |
122 |
379 |
|
*Variant B.1.1.7 was found in only two patients (oral and cervical cancer) and was not included in this table.
DISCUSSION
Genomic surveillance is essential to monitoring the SARS-CoV-2 variants in circulation in different populations and at distinct geographic locations, and ultimately measuring their impact on public health. In this study, the Sanger sequencing was used to characterize the SARS-CoV-2 variants infecting 384 cancer patients, based on the service of COVID-19 diagnosis by RT-qPCR provided by INCA during the crucial period of the pandemic.
It has been pivotal to assess SARS-CoV-2 dynamics among cancer patients due to their immunosuppressive status. It has been recently suggested by the authors and others that the continuous virus replication and prolonged virus shedding in the immunocompromised host, likely resulted from the inability of an impaired immune system to control virus replication, are thought to play a major role in the emergence of SARS-CoV-2 variants in human populations, and likely even the VOCs that spread and predominate across the COVID-19 pandemic history11,18.
The frequency of the different SARS-CoV-2 variants throughout the timeframe studied (October 2020 to September 2022) parallels the pattern seen in the country as a whole as reported by Fiocruz17 for the same period. Therefore, no specific changes in time with respect to circulation of VOCs were observed among cancer patients and the general Brazilian population. In March and April of 2022 no COVID-19 cases were detected nor samples were collected, but these were the months of that year where COVID-19 cases dropped momentarily overall in the country, between two COVID-19 epidemiological waves. National genomic surveillance data from Fiocruz17 reported the emergence of Omicron subvariant BA.2 in this same period. Of note, this subvariant was detected in the study samples in the subsequent months (May and June 2022). Nevertheless, further analyses did not discriminate among Omicron subvariants, neutralizing any bias that this specific subvariant may have caused.
Since a number of different malignancies are oversaw at INCA, the spectrum of malignancies among COVID-19 patients were compared across the timeframe investigated, and the profiles of cancer topographies were also compared with concomitant SARS-CoV-2 infections. It is well known that patients with hematological malignancies as lymphoma, leukemia and multiple myeloma are more prone to SARS-CoV-2 infection and disease severity than patients with solid tumors due to the impairment of their immune system19,20, but the impact of specific SARS-CoV-2 variants on specific cancer topographies has not been investigated so far.
In an overall analysis comparing the cancer topographies associated with infection by each SARS-CoV-2 variant, the profiles were significantly distinct, i.e. each VOC appears to infect hosts with preferential types of cancer. When looking into specific VOC pairwise comparisons (each VOC against the remaining), some particular associations were found, such as skin cancer with Gamma and P.2 infection, breast and cervical cancer with Gamma infection and rectum and prostate cancer with Omicron. Although these data are preliminary and observational, they suggest that patients with specific types of cancer appear more susceptible to particular SARS-CoV-2 variants. To the best of the existing knowledge, this is the first documentation of such association in the literature.
The present study has several limitations. The small number of samples and the reduced number of samples per month could compromise the interpretation of the results. However, the patterns of viral variants circulating in each month parallels those reported countrywide, with large number of samples. Importantly, there was a reduced income of samples for testing from INCA’s Hospital do Câncer II and III, which are in charge of managing gynecological and breast cancer patients, respectively, after November 2021. This fact certainly impacted the low number of breast (n = 2) and cervical (n = 1) samples among Omicron-infected patients (see Table 3). On the other hand, the additional associations observed herein (e.g. between P.2 or Gamma and skin cancer and between Omicron and rectum and prostate cancer) are still noteworthy.
CONCLUSION
A two-year history of SARS-CoV-2 genomic surveillance at the most important cancer reference center of Brazil has been portrayed. Infections followed the overall viral wave patterns of the country across the period investigated, but interesting insights on associations between specific viral lineages and cancer topographies have been provided, which require further research. The study data contributed to an in-depth understanding of SARS-CoV-2 infection biology in immunosuppressed subjects as cancer patients.
ACKNOWLEDGEMENTS
To the members of INCA COVID-19 Task Force, the clinical staff and patients from the INCA whose cooperation provided conditions and samples that enabled the execution of this study. To Dr. Renata Olício for providing assistance with Sanger DNA sequencing. To GISAID Database (https://www.gisaid.org/), the authors and laboratories for the SARS-CoV-2 genomes data shared.
CONTRIBUTIONS
Marcelo Alves Soares, Juliana Domett Siqueira and Livia Ramos Goes designed the study, wrote, revised and edited the article, and provided expert advice on experimental planning and data interpretation. Élida Mendes de Oliveira, Caroline Carvalho de Sá, Julia Botto de Barros Cordeiro wrote the article. Élida Mendes de Oliveira, Caroline Carvalho de Sá, Julia Botto de Barros Cordeiro, Maria Eduarda Lanzillota Assumpção, Gabriella Seara de Andrade and Vinicius Figueiredo Vizzoni optimized all reagents, performed the sequencing and analysis and collected the clinical data. Luiz Claudio Santos Thuler performed all the statistical analysis, revised and edited the article. João Paulo de Biaso Viola provided expert advice on experimental planning and data interpretation. All the authors approved the final version to be published.
DECLARATION OF CONFLICT OF INTERESTS
There is no conflict of interest to declare.
FUNDING SOURCES
This work was supported by the Brazilian Research Council (305765/2015-9; 307042/2017-0), Carlos Chagas Filho Rio de Janeiro State Science Foundation (E-26/202.894/2017; 202.640/2019; 211.562/2019 and 010.000162/2020). Livia Ramos Goes, Caroline Carvalho de Sá and Julia Botto de Barros Cordeiro were the recipients of a grant from Carlos Chagas Filho Rio de Janeiro State Science Foundation (E-26/200.584/2022, E-26/202.074/2022, E-26/202.078/2022). Juliana Domett Siqueira have received postdoctoral fellowship and Élida Mendes de Oliveira, Maria Eduarda Lanzillota Assumpção and Gabriella Seara de Andrade received undergraduate students fellowships by the Brazilian Ministry of Health while conducting this study.
REFERENCES
1. World Health Organization [Internet]. Geneva: 2020. Coronavirus disease (COVID-19) pandemic Overview. [accessed 2024 Mar 19]. Available at: https://www.who.int/europe/emergencies/situations/covid-19
2. Scovino AM, Dahab EC, Vieira GF, et al. SARS-CoV-2's variants of concern: a brief characterization. Front Immunol. 2022;13:834098. doi: https://doi.org/10.3389/fimmu.2022.834098
3. Centers for Disease Control and Prevention. SARS-CoV-2 Variant classifications and definitions. [Internet]. [Atlanta]: CDC; 2023. [update 2023 Sep 1; accessed 2024 Mar 19] Available at: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html
4. Pinheiro JR, Reis EC, Farias JP, et al. Impact of early pandemic SARS-CoV-2 lineages replacement with the variant of concern P.1 (Gamma) in Western Bahia, Brazil. Viruses. 2022;14(10):2314. doi: https://doi.org/10.3390/v14102314
5. World Health Organization [Internet]. Geneva: WHO; 2021. COVID-19: Surveillance, case investigation and epidemiological protocols - Guidance for surveillance of SARS-CoV-2 variants: Interim guidance, 9 August 2021, 2021 ago 9. [update 2023 Sep 1; accessed 2024 Mar 19] Available at: https://www.who.int/publications/i/item/WHO_2019-nCoV_surveillance_variants
6. Ren SY, Wang WB, Gao RD, et al. Omicron variant (B.1.1.529) of SARS-CoV-2: Mutation, infectivity, transmission, and vaccine resistance. World J Clin Cases. 2022;10(1):1-11. doi: https://doi.org/10.12998/wjcc.v10.i1.1
7. Chakraborty C, Bhattacharya M, Sharma AR, et al. A comprehensive analysis of the mutational landscape of the newly emerging Omicron (B.1.1.529) variant and comparison of mutations with VOCs and VOIs. Geroscience. 2022;44(5):2393-425. doi: https://doi.org/10.1007/s11357-022-00631-2
8. Mlcochova P, Kemp SA, Dhar MS, et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599(7883):114-9. doi: https://doi.org/10.1038/s41586-021-03944-y
9. Lee ARYB, Wong SY, Chai LYA, et al. Efficacy of covid-19 vaccines in immunocompromised patients: systematic review and meta-analysis. BMJ. 2022;376:e068632. doi: https://doi.org/10.1136/bmj-2021-068632
10. Siqueira JD, Goes LR, Alves BM, et al. SARS-CoV-2 genomic analyses in cancer patients reveal elevated intrahost genetic diversity. Virus Evol. 2021;7(1):veab013. doi: https://doi.org/10.1093/ve/veab013
11. Goes LR, Siqueira JD, Garrido MM, et al. Evidence of recurrent selection of mutations commonly found in SARS-CoV-2 variants of concern in viruses infecting immunocompromised patients. Front Microbiol. 2022;13:946549. doi: https://doi.org/10.3389/fmicb.2022.946549
12. Artic Network [Internet]. SARS-CoV-2 Amplicon Set Version 3. [place unknown]: Artic Network; 2019. [update 2023 Sep 1; accessed 2024 Mar 19]. Available at: https://artic.network/ncov-2019
13. Goes LR, Siqueira JD, Garrido MM, et al. New infections by SARS-CoV-2 variants of concern after natural infections and post-vaccination in Rio de Janeiro, Brazil. Infect Genet Evol. 2021;94:104998. doi: https://doi.org/10.1016/j.meegid.2021.104998
14. Nguyen LT, Schmidt HA, von Haeseler A, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268-74. doi: https://doi.org/10.1093/molbev/msu300
15. Kalyaanamoorthy S, Minh BQ, Wong TKF, et al. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14(6):587-9. doi: https://doi.org/10.1038/nmeth.4285
16. Conselho Nacional de Saúde (BR). Resolução n° 466, de 12 de dezembro de 2012. Aprova as diretrizes e normas regulamentadoras de pesquisas envolvendo seres humanos. Diário Oficial da União, Brasília, DF. 2013 jun 13; Seção I:59.
17. Dashboard Genomic Network [Internet]. Rio de Janeiro: Fiocruz; 2024. SARS-CoV-2 Genomic Surveillance in Brazil, 2024 jul 9. [update 2023 Sep 1; accessed 2024 Mar 19] Available at: https://www.genomahcov.fiocruz.br/dashboard-en/
18. Markarian NM, Galli G, Patel D, et al. Identifying markers of emerging SARS-CoV-2 variants in patients with secondary immunodeficiency. Front Microbiol. 2022;13:933983. doi: https://doi.org/10.3389/fmicb.2022.933983
19. Costa GJ, Azevedo CRAS, Júnior JIC, et al. Higher severity and risk of in-hospital mortality for COVID-19 patients with cancer during the year 2020 in Brazil: A countrywide analysis of secondary data. Cancer. 2021;127(22):4240-8. doi: https://doi.org/10.1002/cncr.33832
20. Silva JL, Souza BSW, Albuquerque LZ, et al. Factors influencing COVID-19 mortality among cancer patients: a brazilian multi-institutional study. PLoS One. 2023;18(12):e0295597. doi: https://doi.org/10.1371/journal.pone.0295597
Recebido em 1/4/2024
Aprovado em 8/7/2024
Scientific-editor: Anke Bergmann. Orcid iD: https://orcid.org/0000-0002-1972-8777
![]()
Este é um artigo publicado em acesso aberto (Open Access) sob a licença Creative Commons Attribution, que permite uso, distribuição e reprodução em qualquer meio, sem restrições, desde que o trabalho original seja corretamente citado.